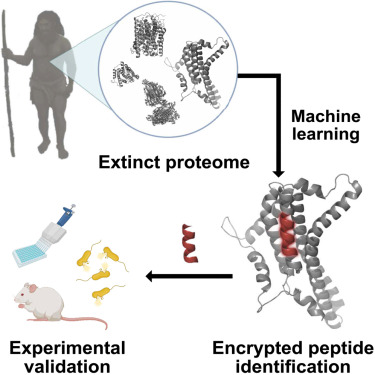
In a groundbreaking study, bioengineers have harnessed the power of artificial intelligence (AI) to bring back extinct molecules, offering new possibilities for antibiotic development. By applying computational methods to data from modern humans and our extinct relatives, Neanderthals and Denisovans, researchers have identified molecules that could combat bacteria and inspire the creation of new drugs to treat human infections.
The Need for New Antibiotics
Over the past few decades, the development of antibiotics has stagnated, with most prescribed medications on the market for over 30 years. In the meantime, antibiotic-resistant bacteria are on the rise, necessitating the development of new treatments. The search for effective antibiotics has become a global concern.
Unlocking the Potential of Extinct Species
Many organisms produce short protein subunits called peptides, which possess antimicrobial properties. While some of these peptides, derived from bacteria, are already in clinical use, the proteins from extinct species remain an untapped resource for antibiotic development.
From Jurassic Park to Molecular Resurrection
Inspired by the movie Jurassic Park, where scientists revived dinosaurs, the research team took a more feasible approach: why not bring back molecules? They trained an AI algorithm to recognize specific sites on human proteins where these proteins are cut into peptides. By applying this algorithm to protein sequences of modern humans, Neanderthals, and Denisovans, the researchers identified new peptides that could potentially kill bacteria.
Accelerating the Search for Antibiotics
Utilizing AI for peptide discovery significantly accelerated the process. While it typically takes three to six years using traditional methods to discover a single new antibiotic, AI-based peptide identification takes a matter of weeks. This efficiency could revolutionize drug discovery for fighting bacterial infections.
Testing the Power of Extinct Molecules
The researchers tested dozens of peptides in laboratory dishes to determine their effectiveness in killing bacteria. From these tests, they selected six powerful peptides derived from modern humans, Neanderthals, and Denisovans. When administered to mice infected with Acinetobacter baumannii, a common cause of hospital-borne infections, all six peptides halted the growth of the bacteria in thigh muscles, but none were able to completely eliminate the infection. However, five of the molecules did successfully kill bacteria in skin abscesses, albeit with high doses.
Potential for Improvement and Future Research
The study’s co-author, Cesar de la Fuente, believes that refining the molecules and algorithms used could lead to more effective versions. Furthermore, he sees the AI-based approach to drug discovery as a promising new avenue for research and development. However, other experts caution that the algorithm needs improvement before molecular de-extinction can have a significant impact on antibiotic development.
New Approach Generates Excitement
Euan Ashley, a genomics and precision-health expert at Stanford University, expressed enthusiasm for this innovative approach in the field of antibiotic development. He was convinced by the team’s research that exploring the ancient human genome could yield valuable insights and potential breakthroughs.
References
Maasch, J. R. M. A., Torres, M. D. T., Melo, M. C. R. & de la Fuente-Nunez, C. Cell Host & Microbe https://doi.org/10.1016/j.chom.2023.07.001 (2023)